SNP Microarray for Products of Conception (POC) Test
Why miscarriages happen?
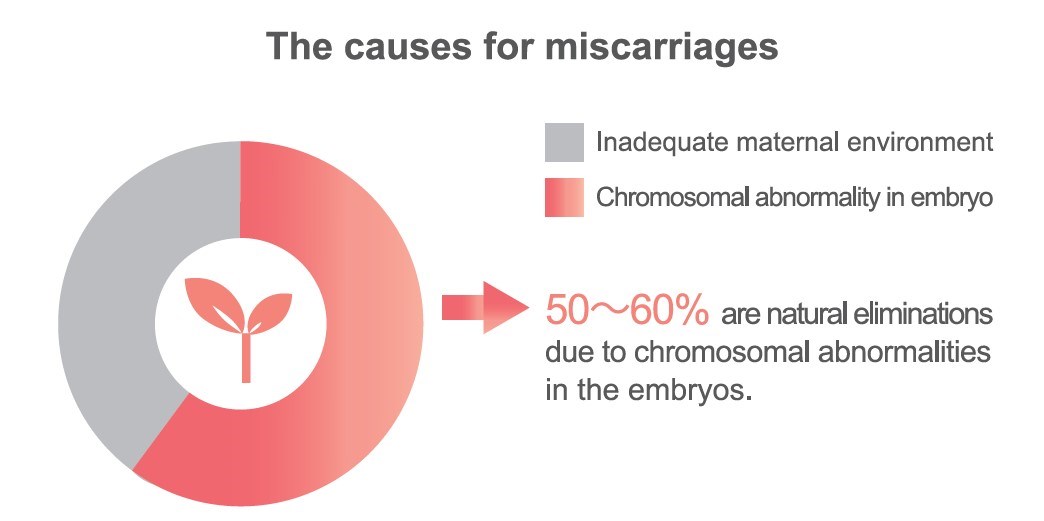
Studies have shown that there is a 15-20% chance of miscarriage during pregnancy, and 5% of couples may have recurrent miscarriages. In addition, 50-60% of the causes for miscarriage is due to chromosomal abnormality in the fertilized egg and is naturally eliminated.
Reference:
1. Stock S: Miscarriage, National Institute of Health; (2012) Retrieved 2013-09-01.
2. Stephenson M, Kutteh W. Evaluation and management of recurrent early pregnancy loss. Clin Obstet Gynecol (2007) 50: 132-145
3. Rai R, Regan L, Recurrent miscarriage. Lancet (2006) 368:601-11
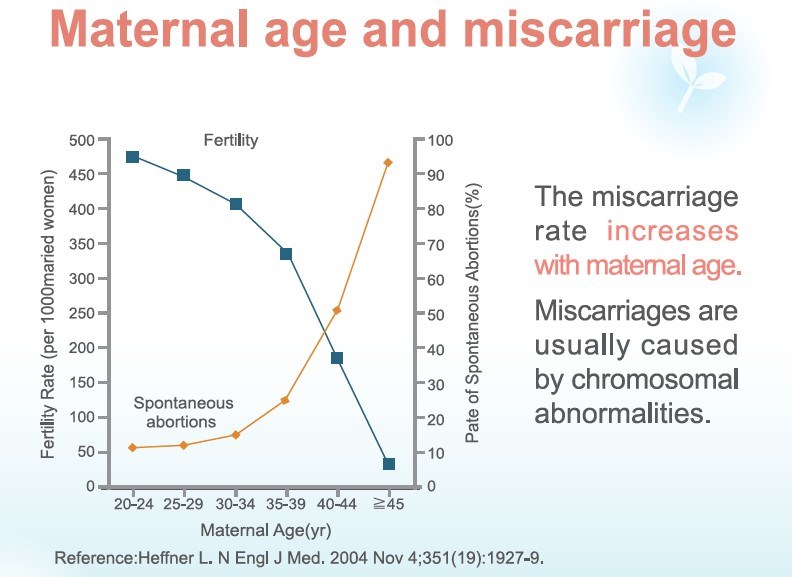
SNP Microarray is recommend in POC testing

- In 2016, the American College of Obstetricians and Gynecologists (ACOG) and the Society for Maternal-Fetal Medicine (SMFM) issued that chromosomal microarray analysis typically can replace the need for fetal karyotype.
- 1. Most genetic changes identified by chromosomal microarray:
- 2. Too small to be identified by standard karyotyping
- 3. NOT associated with increasing maternal age
- 4. Chromosomal microarray analysis is recommended:
- If the fetus is structurally abnormal
- Evaluation of intrauterine fetal death or stillbirth, because of the test’s increased likelihood of obtaining results and improved detection of causative abnormalities
- 5. SNP-based chromosomal microarray of products of conception:
- Yields a higher rate of results compared with karyotyping
- Can identify maternal cell contamination to decreasing false-negative results
- 6. Comprehensive pretest and posttest genetic counseling regarding the benefits, limitations, and results of chromosomal microarray analysis is essential.

- Conclusion of Genetics in Medicine 2017 research:
- 1. Analysis of POC specimens by karyotyping fails in 20-40% of cases. SNP-based chromosomal microarray is a robust platform, with successful results obtained in >90% of cases.
- 2. SNP-based chromosomal microarray can identify aneuploidy, polyploidy, whole-genome homozygosity, segmental genomic imbalances, and maternal cell contamination, thus maximizing sensitivity and decreasing false-negative results.
- 3. Understanding the etiology of fetal loss enables clarification of recurrence risk and assists in determining appropriate management for future family planning.

Why choose GGA SNP Microarray
GGA SNP Microarray with its superior sensitivity and resolution is a more EFFICIENTLY testing to detect more small variation across the whole chromosome set and provide COMPREHENSIVE genetic counseling service.
- Higher detection rate than karyotyping and array-based comparative genomic hybridization (array-CGH)
- More Genetic variants are detected: triploidy, loss of heterozygosity (LOH), or uniparental isodisomy (iso-UPD)
- Avoid test failure related to cell culture, which can occur with pregnancy loss tissues
- Decreasing false-negative results: can identify maternal cell contamination
Specimen requirement and notice
- Placenta (including 50mg chorionic villi) > 1cm3
- Umbilical cord > 1cm
- Fetal Tissue (excluding whole fetus)
- Blood(if follow-up parental testing needed) 3-5mL in Lavender top tube (EDTA)
- Place in sterile tube(s) without any medium, but sterile Hank's balanced salt solution, Ringer's solution, or normal saline are acceptable
- Unacceptable Specimens:whole fetus and specimens that have been fixed by formalin, formaldehyde and alcohol
- Turnaround time (TAT): 20 working days from receiving specimen
Future pregnancy?
If a chromosomal abnormality has been confirmed to be the cause for miscarriage, a discussion with your physicians regarding reproductive implication for future pregnancies can be helpful.
Pre-implantation Genetic Screening (PGS) is a test to screen in vitro fertilized embryos for normal chromosome numbers for transfer selection. Using PGS can effectively increase the rate of pregnancy and live birth for pregnant women of advanced ages.
→To know more about PGS




.jpg)